Image Modulation Profiles
Kim Tolbert and Richard Schwartz (Last update
14-Jan-2022)
On May 3, 2010, routines to compare the expected modulation profiles from
an image to the observed count rates were added to the RHESSI image object
software. Previously, this comparison was plotted and some
parameters stored in the Forward Fit and Pixon algorithms, but now it
can be done for any image algorithm and statistics are saved for separate
detectors. New info parameters containing the result of the
comparison are stored in the image object, and plots can be generated to
show the profiles.
These parameters and plots can be generated during and after construction
of a single image using any RHESSI image algorithm (visibility-based
or non-visiblity-based). However, because the calibrated event
list used to make the image must be available to compute the profiles, for
image cubes we have the following restrictions:
- For image cubes using non-visibility algorithms, the profiles can be
computed for each image as it is made, but not after the image cube is
completed (only the final image calibrated event list is available at
that point.
- For image cubes using visibility algorithms, the profiles can NOT be
computed at all. This is because visibilities for all times and
energies in the cube are computed at once, so the calibrated event list
is not available for each image as it is made.
Using this tool, we've discovered that the CLEAN images often fail to
match the observations when the finer detectors are included. Please
see the section at the end of this page on a new
Clean Regress method for computing the final CLEAN image.
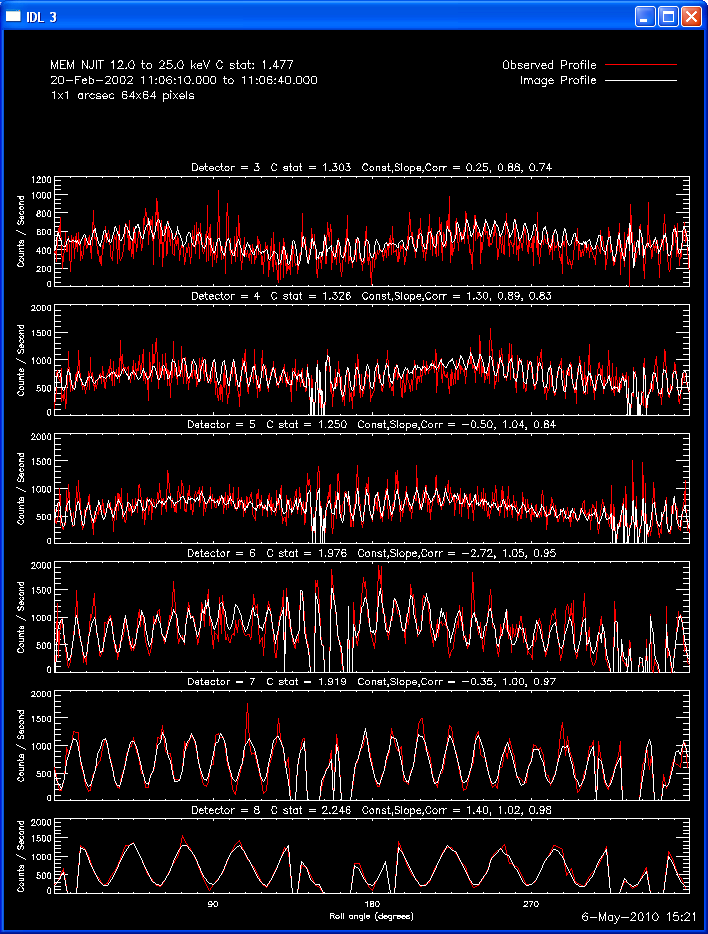
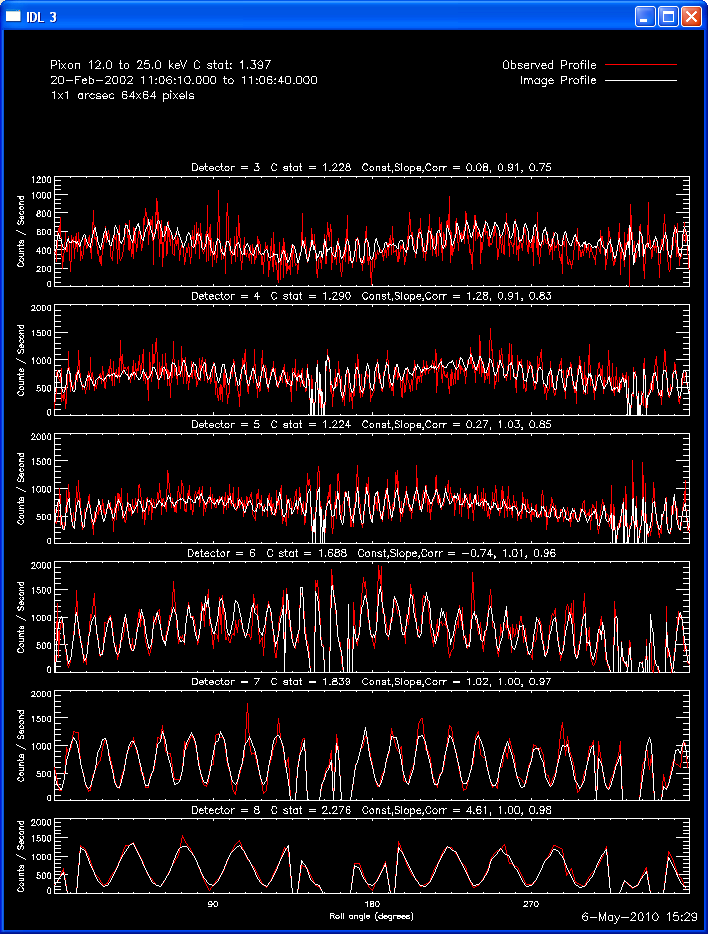
Profile Plots:
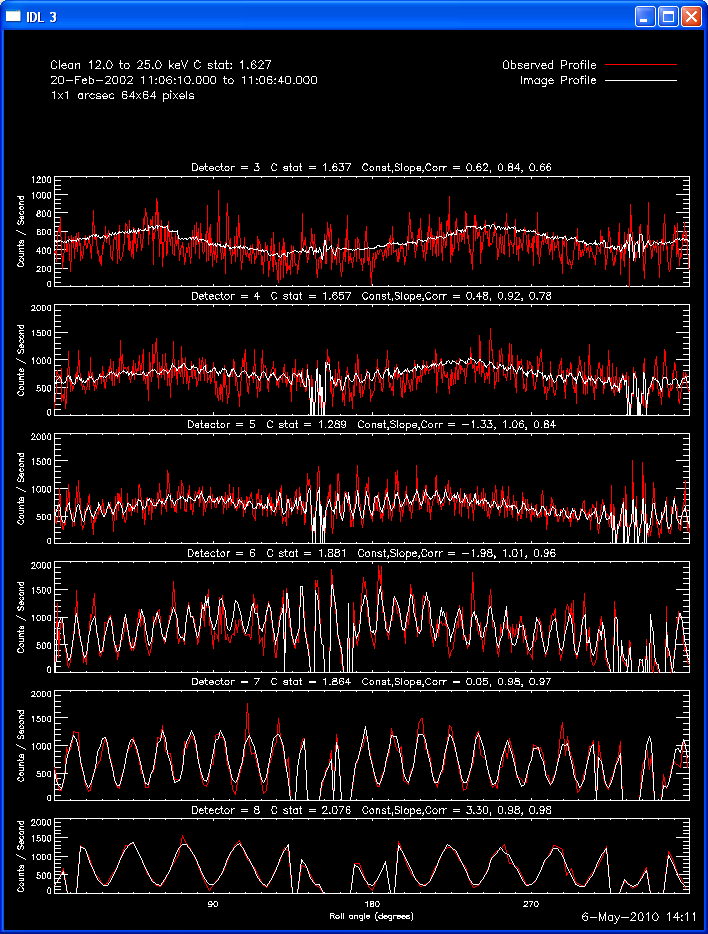
The profile plots show, for each detector selected, the observed
modulation profile in red, overlaid by the expected modulation profile
computed from the image in white. If the phase stacker option was
enabled, the x axis is the roll angle in degrees (each detector's offset
is accounted for so that in the stacked plots, the profiles for each
detector are on the same x axis). If the phase stacker was not
enabled, the x axis is the time of the observation in seconds relative to
the start of the observation. The default plots show Counts /
Second, but Counts / Bin plots can be requested. A residual plot (
(observed - expected )/ sqrt(expected) ) can also be requested.
New Info Parameters:
The following new info parameters are stored for each image for all
algorithms (except vis algorithms in an image cube).
| Name (replace 'xxx' by the prefix for the algorithm
in use, e.g. 'clean') |
Type |
Description |
| xxx_profile_tot_cstat |
Scalar |
The overall C-statistic from the comparison of
expected to observed modulation profiles for all selected detectors |
| xxx_profile_cstat |
Array [9] |
The C-statistic from the comparison of each separate
detector's expected and observed modulation profile. |
| xxx_profile_coeff |
Array [3,9] |
The constant, slope, and correlation coefficient
between the expected profile and observed profile for each detector.
Apply the constant and slope to the expected profile to match the
observed profile. Note that the constant is in units of counts /
bin. The correlation coefficient gives an overall estimate of
the correlation between the expected and observed profiles. |
The C-statistic is computed according to Ed Schmahl's derivation from the
Cash paper (ApJ, 228, 939, 1979) (using the routine
hsi_pixon_gof_calc.pro) which is what Forward Fit and Pixon have been
using.
Retrieve these parameters as you would any other info parameter, e.g. for
a pixon image:
cstat = o->get(/pixon_profile_cstat)
help, o->get(/pixon_profile), /st
How to display profiles from the command line:
These image object control parameters control plotting profiles during
the generation of images:
profile_show_plot - Disable/enable profile plots. Default
is 0.
profile_plot_rate - 0 means plot counts/bin; 1 means plot
counts/sec. Default is 1.
profile_plot_resid - If set, plot profile residuals. Default is 0.
profile_window - Internal variable to keep track of window to plot
profiles in
profile_ps_plot - If set, send plot to PS file (auto-generated name).
Default is 0.
profile_jpeg_plot - If set, send plot to JPEG file (auto-generated name).
Default is 0.
profile_plot_dir - Directory to write PS or JPEG file in. Default is current
directory.
So, for example, if you enter this command (where o is your image
object):
o->set, /profile_show_plot,
/profile_plot_rate
every time you generate an image, a rate profile plot will be displayed
on the screen. If you also set profile_ps_plot, i.e.
o->set, /profile_show_plot,
/profile_plot_rate, /profile_ps_plot
then the plots will be sent to an auto-named PS file in your current
working directory every time you generate an image.
To display the profile later for the most recent image generated, call
the hsi_image_profile_plot routine. See the header of that routine
for all keyword options. Note that options you don't explicitly specify in the keyword arguments will take their value from the corresponding parameter in the image object (i.e. if the image object parameter profile_plot_rate is set to 1, then you must call hsi_image_profile_plot with rate=0 if you don't want a rate plot). Examples (where o is your image object):
hsi_image_profile_plot, o, rate=0 ;
plot profiles in counts/bin on screen
hsi_image_profile_plot, o, /ps
; plot profiles in PS file, units are whatever profile_plot_rate parameter specifies
hsi_image_profile_plot, o,
/rate ; plot profiles in
counts/sec
hsi_image_profile_plot, o,
/resid ; plot residual of
comparison between observed and expected profiles
hsi_image_profile_plot, o, /ps, file_plot='test.ps' ; plot profiles in PS
file called test.ps
You can also retrieve the c-statistic and coefficients, and the values
plotted in the profiles by providing output keyword arguments, for
example:
hsi_image_profile_plot, o, prof_fit=prof_fit,
xp=xp, out_struct=out_struct
How to display profiles from the GUI:
There are two new buttons in the RHESSI image GUI.
To display the profile plot automatically every time you generate an
image, click either the 'Screen' or 'PS' box under 'Profile Plots'.
Tip1: The default plot units are counts/sec, To plot counts/bin
or residuals, type o->set,profile_plot_rate = 0 or
o->set,/profile_plot_resid from the command line.
Tip2: If you want to preserve the last automatically generated profile
window, type o->set,profile_window = -1 from the command line to
force the software to open a new window for the next profile.
If you want to display the profiles after an image was generated,
use the option in the 'Display->' button called 'Count Profile vs
Roll Angle'. There are three options from that button - 'Counts /
bin', 'Counts / sec', and 'Residuals', to allow you to choose what to
plot. These plots are shown in a newly created plot window.
Discussion and Examples
There is one basic objective measure of the success of any image
deconvolution algorithm: it should successfully reproduce the observation
in a way that's consistent with the statistics of the detector measurement
process. In our case, we are counting independent photon events that obey
Poisson statistics. So the predicted counts should agree on average and in
detail with the measurement. These image profiles help us determine this
agreement at a glance. For example, if the predicted profiles are
everywhere larger or smaller than the observed for all detectors then we
know that there is a probably an error in the overall normalization of the
image, certainly it hasn't been optimized. If there is such a discrepancy
in the profile for one detector and not all then there is still a problem
but one that is more subtle. Another way to evaluate the profiles is to
compare the modulation depth of the predicted and observed. The image
source size determines this modulation depth so an image that is too wide
will show insufficient modulation depth to be consistent with the profiles
in one or more of the finer grids. Finally, an exact match between the
predicted and observed profiles is not a requirement or even desirable
with perfect count statistics as simulation has shown that for our images
it means that we will probably have degraded the true image into a
collection of spurious sources.
Points to remember:
- Do the average measured and observed agree? For all detectors? For
some?
- To evaluate modulation use the counts/sec plots
- To evaluate detailed problems, use the residual plots.
- The correlation coefficient is also a good way to evaluate
size-dependent issues.
- Variations in the slope parameter as a function of detector means
something is missing in the model as a function of detector. This might
be a problem in the detector response model or that one detector is more
sensitive to pileup than another.
- There are three ways to display the differences in the predicted and
the observed. The residual plot scaled by sigma shows statistically
significant differences. The rate plot best shows when the modulation is
misrepresented. Finally, the counts plot strikes a balance showing some
of the modulation but leaving in enough information to evaluate the
significance of the differences.
- The meaning of the X axis roll angle has been modified (from other
representations of these profiles) to force all of the grids to have the
same orientation by correcting for the grid offset angle (from
hsi_grid_parameters.pro). So a line source aligned to the N/S axis will
produce its maximum modulation at this origin and 180 degrees.
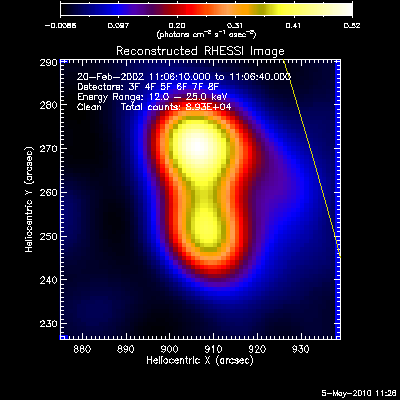
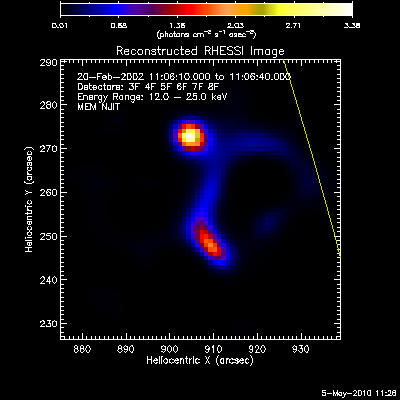
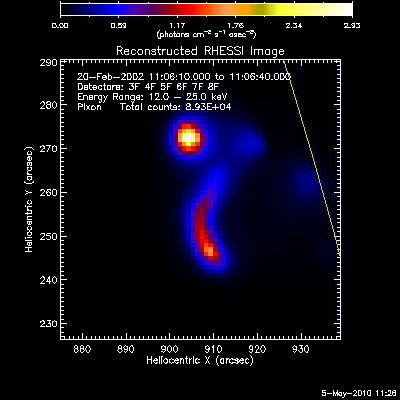
Here are some examples of the profile plot for images of the Feb-20-2002
flare using several algorithms. The time of the image is 20-Feb-2002
11:06:10.000 to 11:06:40.000, phase stacker was enabled, weighting
was natural, and the flux smoothing time was 2. s.
We have noticed problems with the final Clean image of some flares when
we've included the finer grids. The final step of the Clean algorithm as
currently implemented is to add the Clean components map to the residual
map to produce the final map. In certain cases, this results in final
images that fail miserably on Point 1 above - the average measured and
observed do not agree.
We have implemented an alternate method for combining the component and
residual map that will guarantee a better match between the average
observed and measured profiles. This method combines the components map
with the residual map based on a regression of the count rate profiles
from each of the two maps against the observed profile. (Unfortunately,
mismatches caused by problems in single detectors will remain.) The
new method is enabled in the objects by setting the parameter
CLEAN_REGRESS_COMBINE to 1 (via a button in the Clean 'Set Parameters'
widget, or o->set, /clean_regress_combine). The default value of
this parameter is 0, meaning the new method is not used by default.
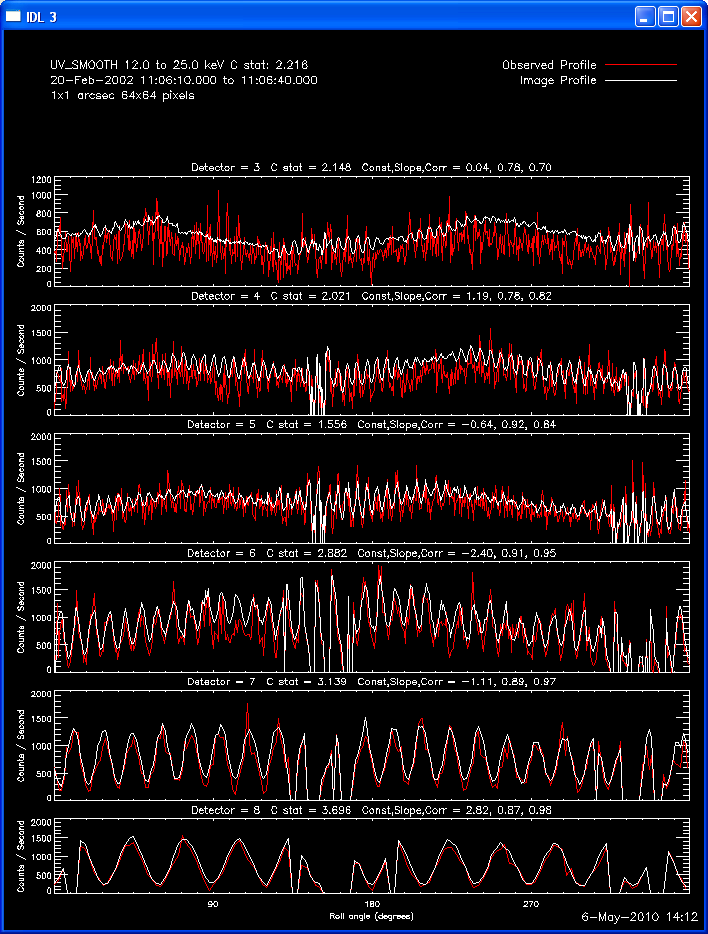
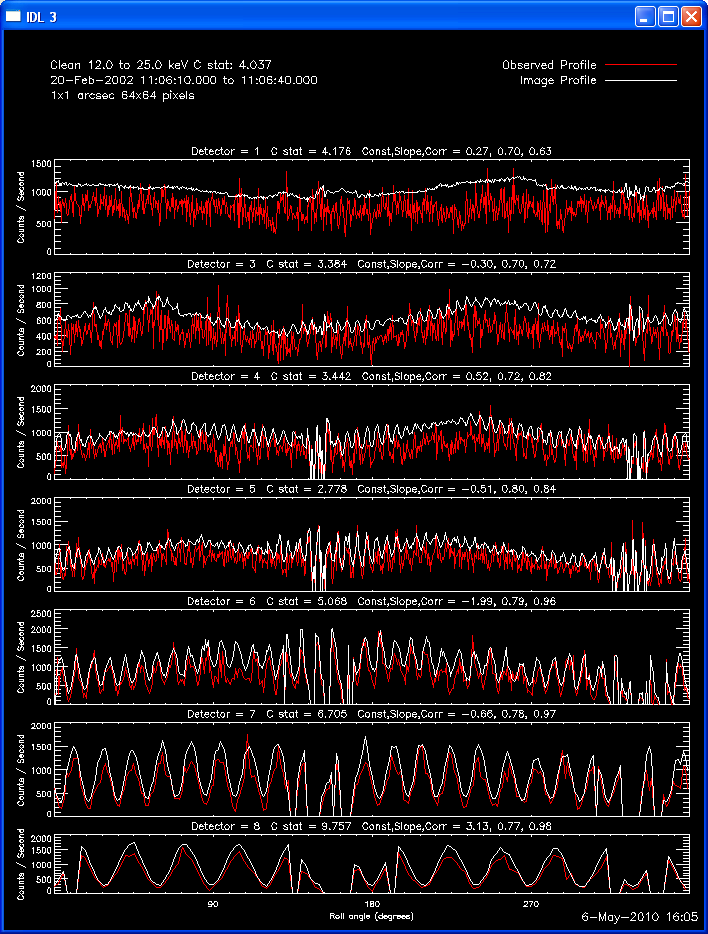
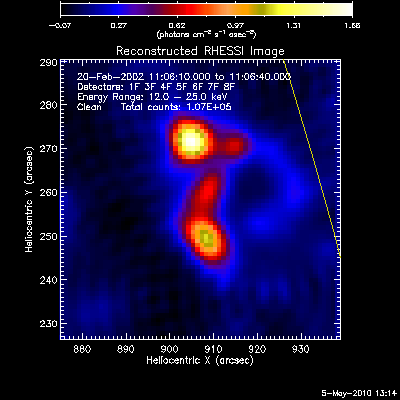
The first image and profile below shows the problematic Clean image for
the same image in the example above, except that Grid 1 was
included. Note the offset between the image and observed rates (this
is reflected in the slopes of ~.7 - .8; a good match has a slope of 1.)
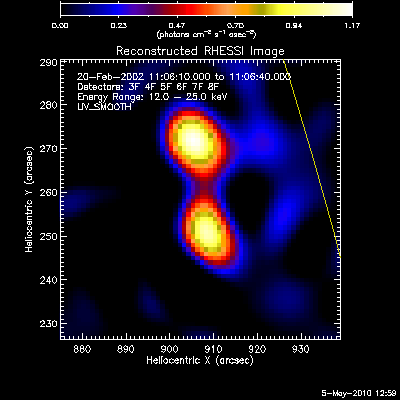
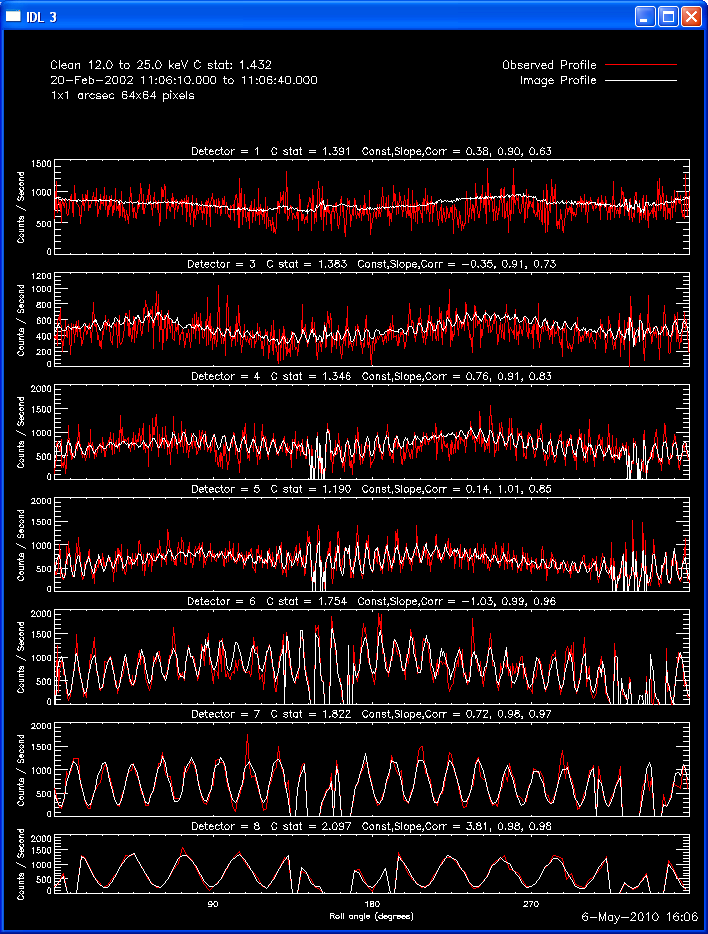
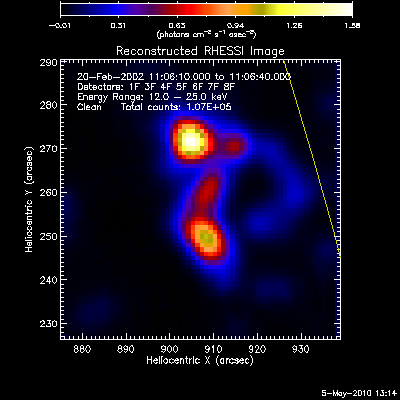
The second image and profile below show the better Clean regressed
image. By using regression, we force the fluxes/profiles to match on
average (note the slopes of .9 - 1.0). This also helps mitigate the
effect of incomplete Cleaning where there may be a patch of flux left in
the residuals. By using regression we preserve some of the inherent
uncertainties from the residuals, but make the best linear combination of
the two maps. Note also that the Clean Regress method does little to
improve the correlation coefficient. Mismatches in source size will barely
be affected by this technique.
An idea for the future: This method suggests a method for selecting
the best convolving beam width to use with the point sources in the
clean_source_map. We could take the component map, convolve it with a
Gaussian of a given width, fit the final result using the Regress method,
and then do an overall comparison for the best C statistic. Since that
would probably favor overly compact source sizes, we could also add a cost
function that would favor smoothness.
Last updated: 12-Jun-2017